Cell motility
Locomotion and movement of living cells
Cell motility is extremely important for many aspects of life from embryonic development and immunity response to wound healing and diseases.
It is not fully understood how cells coordinate overall motility, but from experiments it was established that at least four different stages of locomotion can be distinguished. Cells crawl by extending pseudopodia (filopodia or lamellipodia), adhere, contract and detach the rear. Extension of both filopodia and lamellipodia of most cells is based on actin polymerization. While the protrusive event of cell locomotion is thought to be driven by actin polymerization, the mechanism of forward translocation of the cell body is not completely understood.
| Young's modulus (kdyn/cm 2) | Traction force (kdyn/cm 2) | Cell speed (µm/min) | Projected cell area (10 3 µm 2) |
| 140 | 6.2±1.3 | 0.44±0.14 | 1.74±0.14 |
| 300 | 10.9±3.4 | 0.54±0.13 | 2.18±0.17 |
Table 1. Experimental data from Lo et al., 2000.
It was shown recently that individual fibroblast movement can also be guided by physical interactions at the cell-substrate interface. During experiments conducted in C.M. Lo, H.B. Wang, M. Dembo and Y.L. Wang, Cell movement is guided by rigidity of the substrate, Biophysical J. 79, 144-152, 2000 fibroblasts were cultured on flexible polyacrylamide sheets coated with type I collagen. The rigidity transition was introduced in the center of the sheet, creating "soft'' and "stiff'' sides of substrate with Young's modulus of 140 kdyn/cm 2 and 300 kdyn/cm 2 correspondingly. It was observed that locomoting fibroblasts could easily migrate across the boundary from soft to stiff side. However, cells approaching the boundary from stiff side never crossed it, but continued to crawl on a stiff side along the boundary. It was also shown that spreading area of cells as well as generated traction forces increase on the stiff side of the substratum. Average traction forces generated by the cells on soft side of the substrate were 6.2 kdyn/cm 2, whether traction forces measured on the stiff side were much larger (10 kdyn/cm 2). Average cell speed was also different dependent on substrate rigidity. It was 0.44 µm/min for cells on soft side and 0.54 µm/min for cells crawling on the stiff side. Similar trend was seen for the projected cell area (see Table 1 for summary).
| Substrate rigidity (kdyn/cm 2) | Active force (kdyn/cm 2) | Cell speed (µm/min) | Projected cell area (10 3 µm 2) |
| 140 | 2.7 | 0.43 | 1.65 |
| 300 | 9.1 | 0.69 | 2.28 |
Table 2. Cell model results.
We developed a discrete model (which will be described in the paper we plan to publish soon) of cell motion of a fibroblast on a two-dimensional substrate with a step in rigidity as in experiment (Lo et al., 2000). The cell cytoskeleton is modeled as series of elastic springs and viscous dash-pots in parallel.
Summary for calculated cell speed, projected cell area and active force on different substrate rigidities is shown in Table 2. Values of cell speed and cell area compare well with experimental values (Lo et al., 2000).
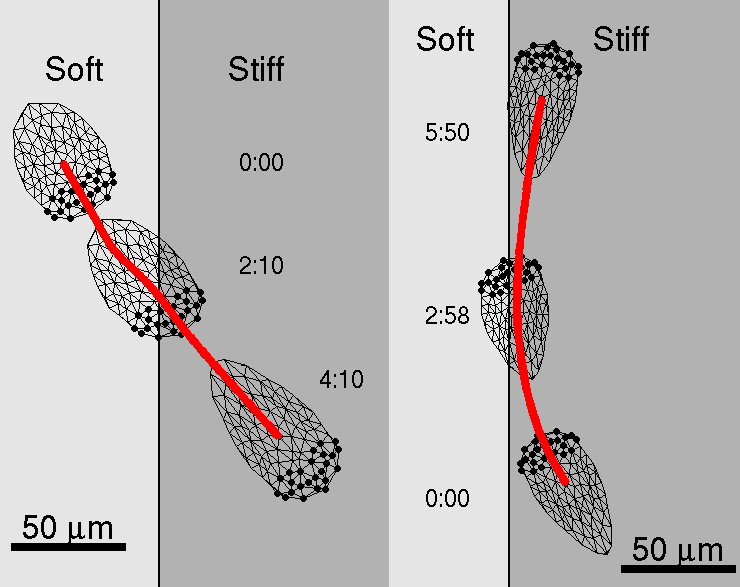
The model cell shows preference for locomotion over stiffer side of the substrate when approaches the boundary between the "soft" and the "stiff" sides of the substrate.
A model cell "planted" on the soft side of the substrate crosses the boundary and continues to move on the stiff side of the substrate. In this case, while cell crosses boundary, its shape changes, speed increases, cell length and projected area grow (see Figures below and Movie "soft-stiff").
A model cell "planted" on the stiff side of the substrate does not cross the boundary between two substrate rigidities, but turns away from the boundary and stays on the stiff side. During the turn at the boundary model cell also experiences changes in shape, speed, length and projected area (see Figures below and Movie "stiff-soft").
This behavior is similar to the experimentally seen (compare with Fig.1 in Lo et al., 2000).
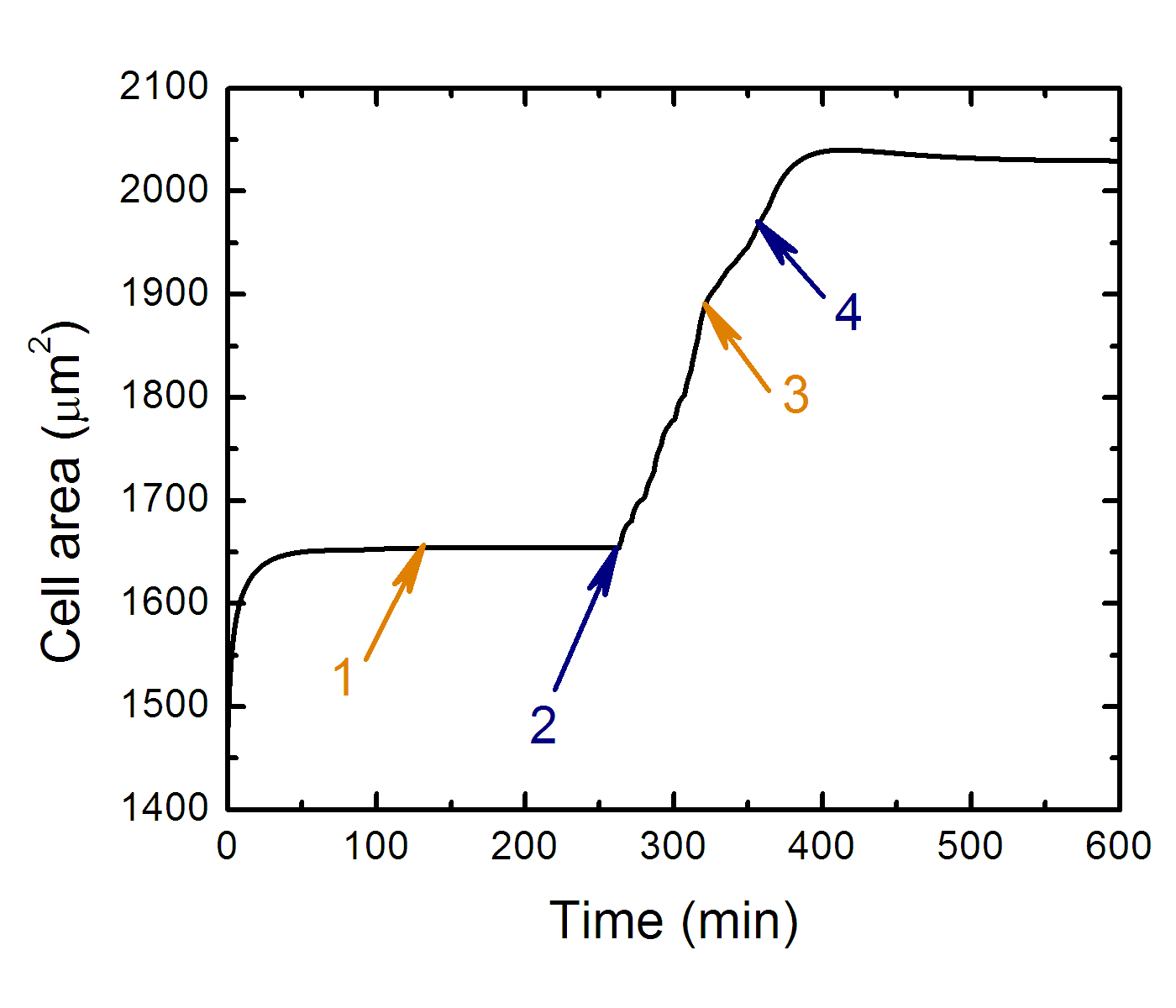
Projected cell area as the function of time along cell trajectory
while it crawls from soft side to stiff side.
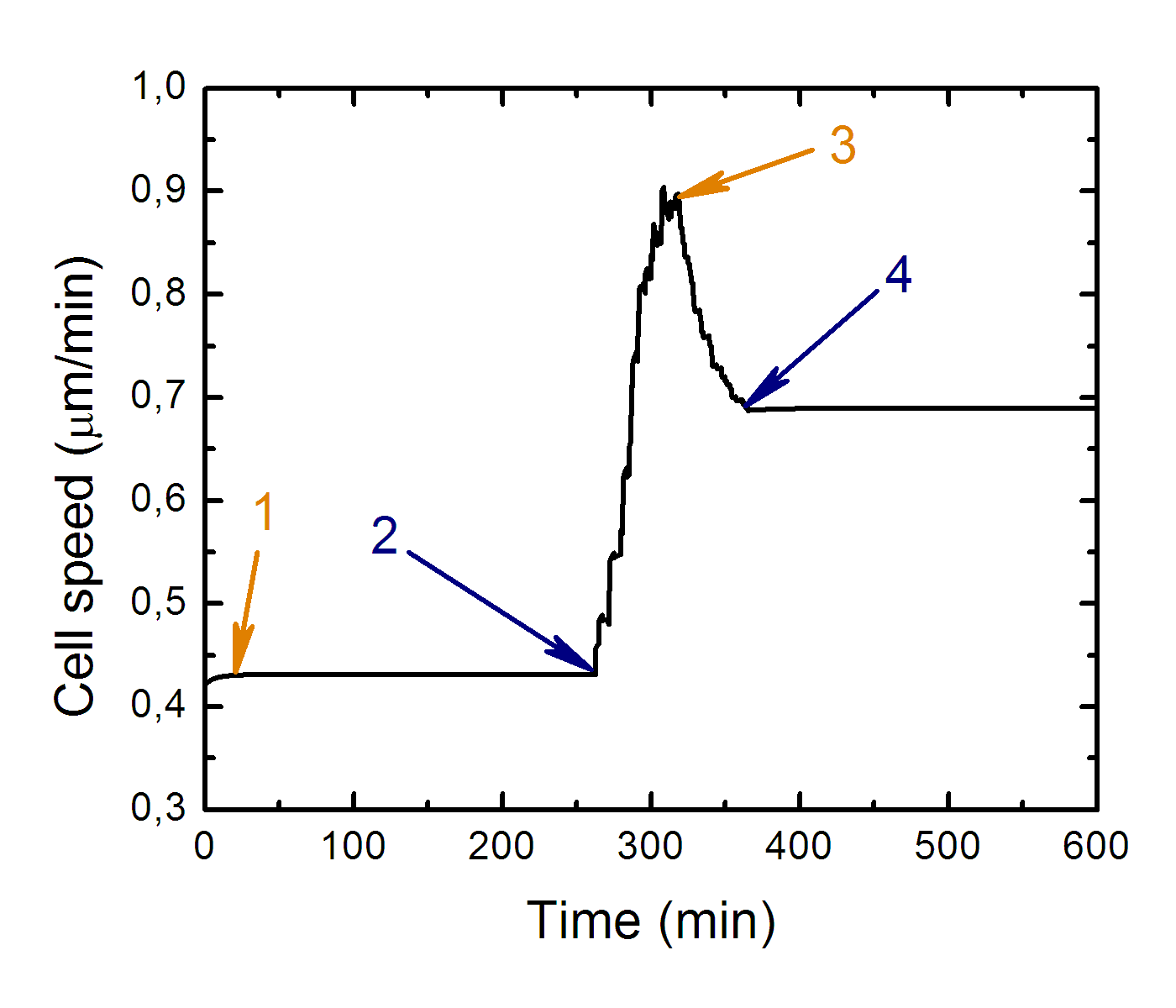
Cell speed as the function of time along cell trajectory while it
crawls from soft side to stiff side.
Where: 1 - transition to equilibrium, 2 - cell contact with "soft-stiff" boundary, 3 - cell front leaves soft side of substrate, 4 - whole cell body leaves soft side of substrate.
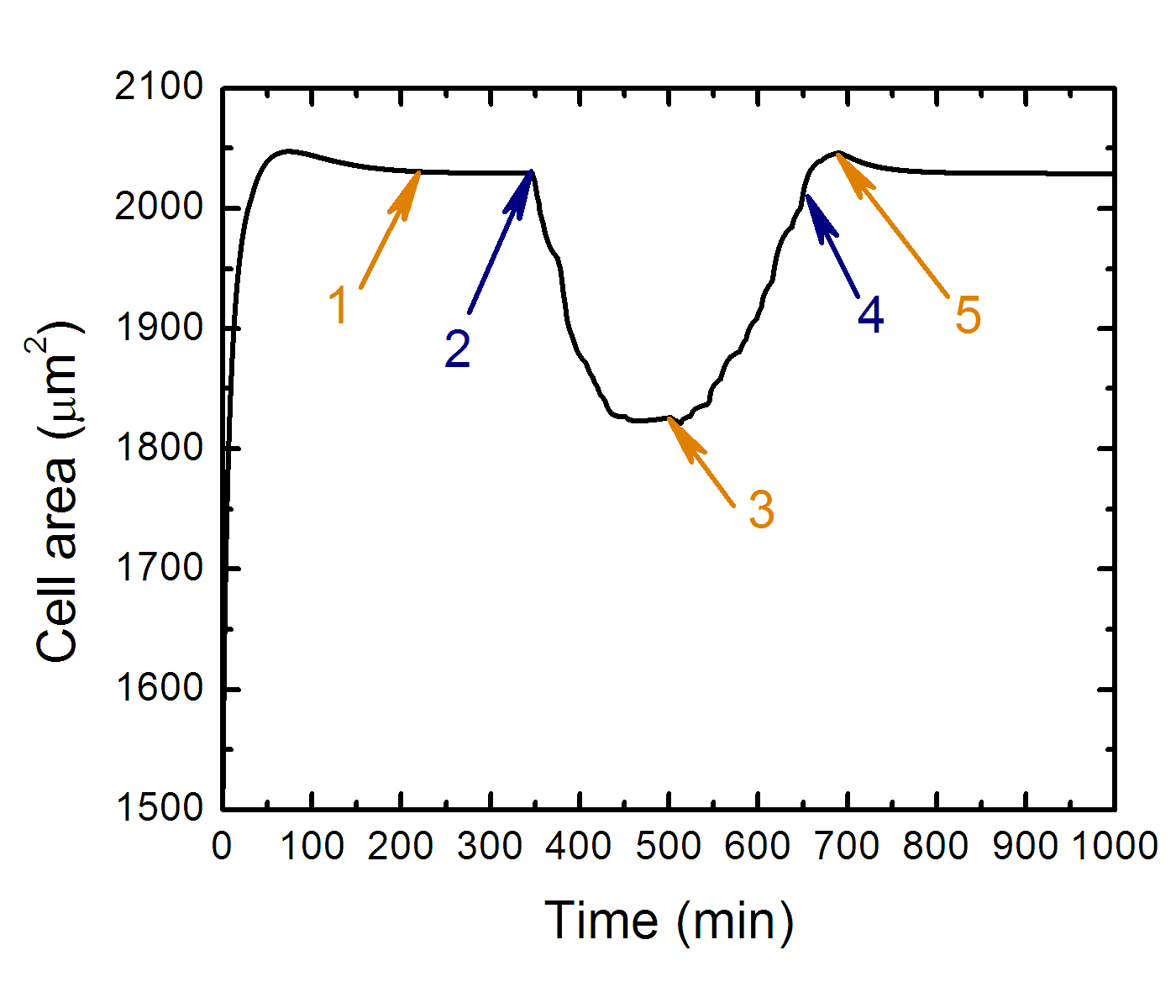
Projected cell area as the function of time along cell trajectory
while it approaches the boundary from stiff side of substrate.
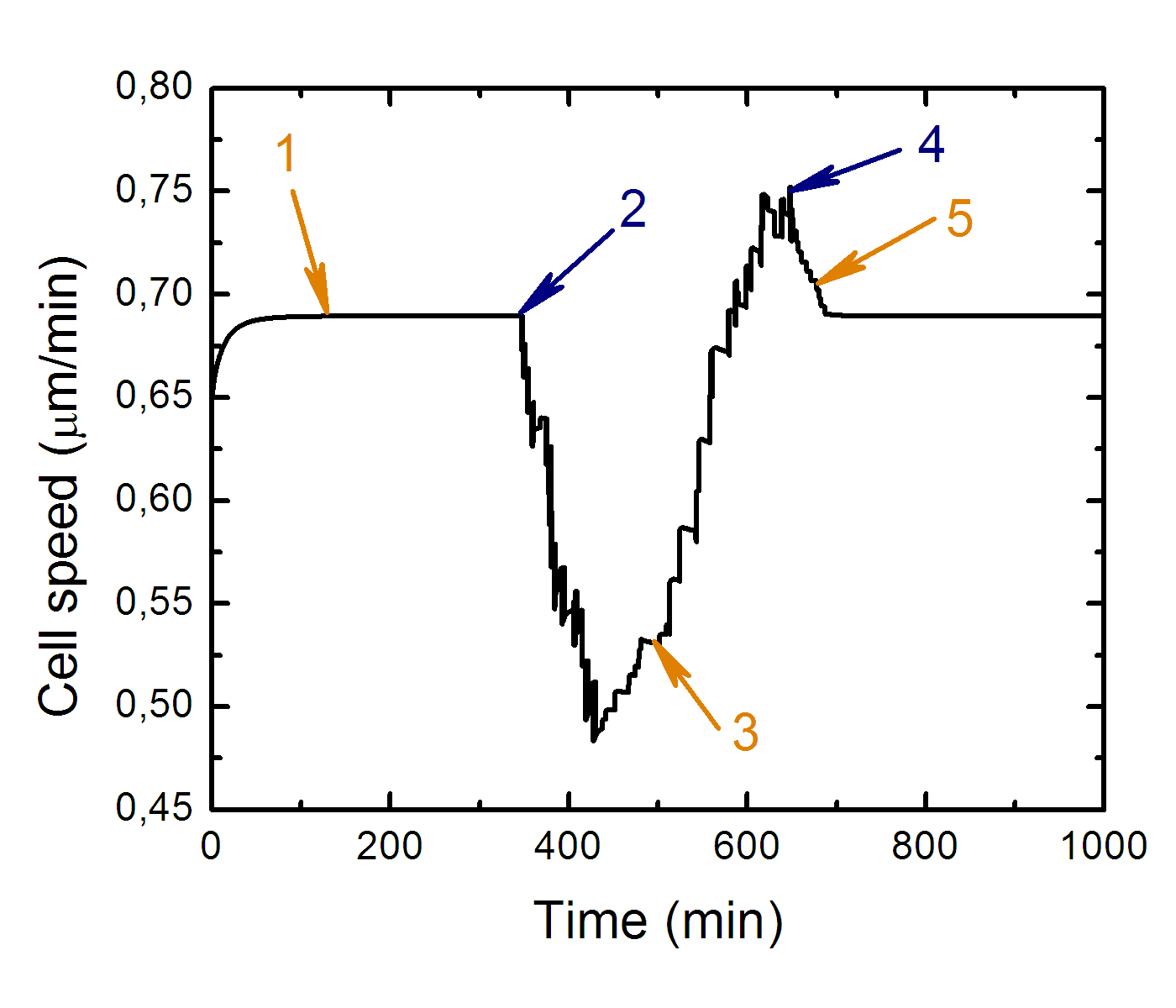
Cell speed as the function of time along cell trajectory while it
approaches the boundary from stiff side of substrate.
Where: 1 - transition to equilibrium, 2 - cell contact with "stiff-soft" boundary, 3 - the cell front over the soft side of substrate at the most, 4 - cell front leaves soft side of substrate, 5 - whole cell body leaves soft side of substrate.
Related papers
- "A model of fibroblast motility on substrates with different rigidities", I. Dokukina, M. Gracheva, Biophysical Journal 98(12), p. 2794-2803 (2010).
- "A continuum model of motility in amoeboid cells", M.E. Gracheva and H.G. Othmer, Bulletin of Mathematical Biology 66, p. 167-193 (2004).