Slowing down and stretching DNA molecule
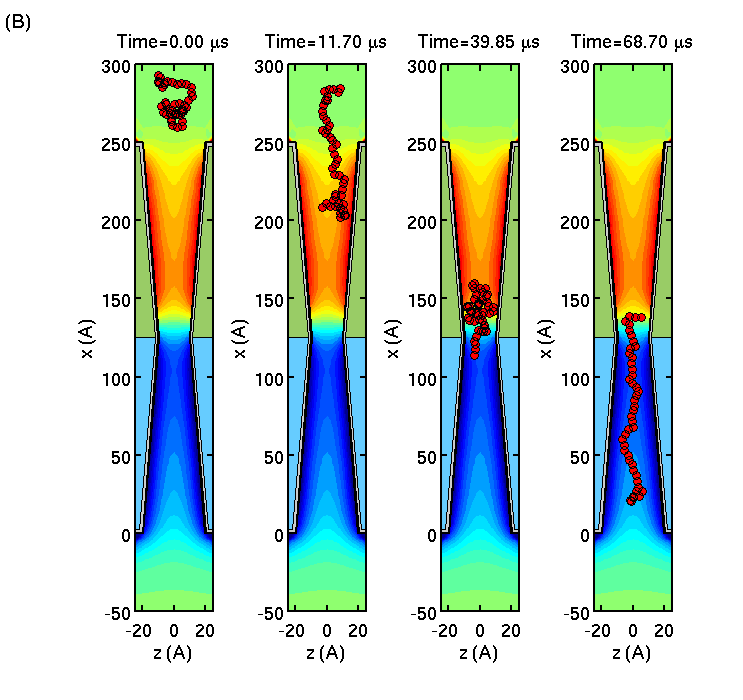
Numerical simulations can provide us with invaluable insight into the microscopic behavior of molecules as they translocate through artificial nanopores. With this in mind, we have developed a computational tool-box that allows us to examine how polymer dynamics will be affected by the electrostatic fields of semiconductor membranes submerged in electrolytic solution.
We consider single-stranded DNA translocation through a semiconductor membrane consisting of doped p- and n-layers of Si forming a pn-junction. Using Brownian dynamics simulations of the biomolecule in the self-consistent membrane-electrolyte potential, we show that the polymer translocation through the membrane is slowed down, while polymer length is greatly extended.
The biomolecule elongation is particularly dramatic on the n-side of the membrane where the lateral membrane electric field restricts (focuses) the biomolecule motion more than on the p-side. The latter effect makes our membrane a solid state analog of the alpha-hemolysin biochannel.
The results indicate that the tunable electric field inside the membrane can effectively control the dynamics of a DNA in the channel to either momentarily trap, slow down or allow the biomolecule to translocate at will, while stretching it.
Related papers
- "Polymer Translocation Through an Electrically Tunable Nanopore in a Multilayered Semiconductor Membrane", Dmitriy V. Melnikov, Alexey Nikolaev, Jean-Pierre Leburton, and Maria E. Gracheva, Chapter in "Nanopore-based technology: single molecule characterization and DNA sequencing", edited by M. E. Gracheva (Humana Press, 2011).
- "Slowing Down and Stretching DNA with an Electrically Tunable Nanopore in a p-n Semiconductor Membrane", Dmitriy V. Melnikov, Jean-Pierre Leburton, and Maria E. Gracheva,Nanotechnology, 23, 255501 (2012).
- "DNA Translocation through a Nanopore in a Single-Layered Doped Semiconductor Membrane", Ining A. Jou, Dmitriy V. Melnikov, Christopher R. McKinney, and Maria E. Gracheva, Physical Review E 86, 061906 (2012).